AI LAB Notes: Adding symbolic AI programming to NVIDIA Jetson powered deep neural network development systems
The following article is written by Dr. Robert B. Trelease from the David Geffen School of Medicine at UCLA and UCLA Brain Research Institute (BRI). We are glad to share here and learn about how current GPUs help scientific research and medicine.
Please also check our previous AI Lab notes about Dr. Trelease working on integrating CLIPS with the tremendous L4T CV and D/CDNN development environment using NVIDIA RTX and Jetson Xavier NX.
?Also don’t miss out latest reComputer Jetson-20 with Jetson Xavier NX production module.
Several decades after its inception, artificial intelligence (AI) development is now driven by widely useful connectionist systems and deep neural network (DNN) simulations that depend on large-scale parallel computing with graphics processing units (GPUs). It is truly the age of high-performance GPU computing, from gaming, 3D simulation, and robotics, through professional design, scientific research, and medicine.
In large part, this AI renaissance has been powered by NVIDIA GPU cards and systems, with their latest Jetson embedded processors providing real-time computer vision, machine learning, natural language, and autonomous machine capabilities in compact low-power systems.
Given their teraOPs GPU processing speed and a solid base of proven DNN software resources, one could ask why ‘Python friendly’ Jetson developers might want to add ‘old traditional’ symbolic programming tools to their JetPack L4T environments.
Simply put, with continually evolving AI methods, newer hybrid neural-symbolic (or neurosymbolic) programming is being increasingly adopted for enhancing the explainability of DNN-based functions, the efficiency of machine learning, and the availability of complementary new methods for inference. https://cacm.acm.org/magazines/2022/4/259402-toward-a-broad-ai/fulltext
Over the past two years, the author has worked on assembling a set of compatible open source symbolic AI programming tools and utility applications that will integrate well with the Jetsons’ Arm64 L4T (Ubuntu) operating system. These include amd64 Ubuntu 18.04 versions for the EGX-class Xeon 20 + RTX GPU workstation that is the Jetsons’ JetPack SDK host and NGC development archive container manager.
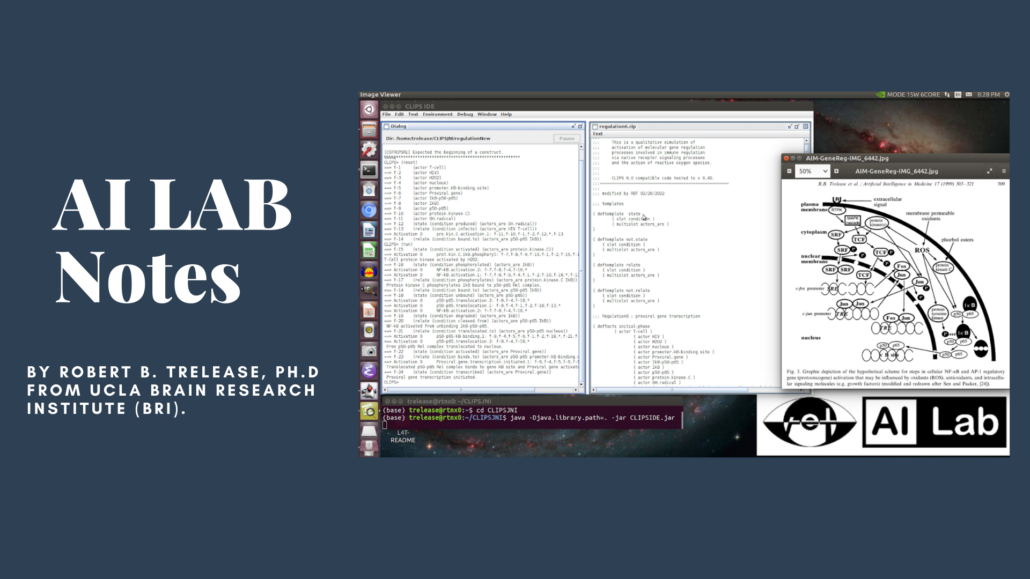
The images below show the first tests of a newly compiled CLIPS 6.40 (2/21) Lisp/Scheme like symbolic programming environment running qualitative process simulations on a Jetson Xavier NX. CLIPS is executing though the OpenJDK Java virtual machine via a .jar app launched from a REPL command line. The required CLIPS Java/C .so function library was compiled for Jetson’s specific arm64 OpenJDK 11 installation.
In the first screen capture image, the upper left bash terminal session shows the end of a successful run of the classic AI ‘monkey and bananas’ cognitive decision-making simulation from test code provided with the CLIPS source.
The lower part of the same terminal screen (below the green/blue command lines) shows loading and running an experiment from the author’s legacy gene regulation process simulation code.
Each line represents a molecular process rule firing report, showing one gene activation ‘pathway’ through the complex interacting ‘web’ of cellular molecular processes responsible for gene regulation. This molecular biology process network is graphically depicted in the upper right diagram.
A detailed account of this simulation’s symbolic knowledge base and related molecular biology qualitative process modeling experiments can be found here: https://www.sciencedirect.com/science/article/pii/S0933365799000214
The second screen capture image (below) demonstrates the CLIPSJNI IDE running another gene regulation experiment simulation in a multi-concept verbose mode, listing the initial conditions for the experimental system (actors etc.) as well as the process rule firings’ logic.
The screen displays CLIPSIDE.jar IDE views of a REPL (command line) window (left) and CLIPS Lisp/Scheme like process rule code text window (right). The background CLIPSIDE window menu bar supports point-click files loading, editing, environment functions, execution commands and debug features.
This experiment uses the same basic CLIPS-format symbolic knowledge base (biological molecules concepts and genetic signaling process rules) as the experiment shown above. This demonstrates that a different set of initial biological conditions results in a different pattern of process activations for a proviral gene.

